Abstract
Introduction: Genetic mutations that occur in blood cancer can be broadly classified into nucleotide sequence mutations including duplications or deletions, fusion gene mutations, and abnormal gene overexpression. Among these genetic mutations, gene fusion not only plays an important role in the development of blood cancers, such as leukemia, but is also important as an essential marker for the diagnosis, risk assessment, optimal treatment selection, and residual lesion detection. Most clinical laboratories use conventional cytogenetic testing, fluorescence in situ hybridization, and multiplex reverse transcriptase-polymerase chain reaction (PCR) to detect fusion gene mutations. However, the currently used methods have great limitations in detecting various gene fusions occurring in blood cancer. To overcome these shortcomings, RNA sequencing (RNA-seq) methods are being introduced in clinical laboratories. The authors developed a targeted RNA-seq panel, a bioinformatics pipeline used for targeted RNA-seq to detect gene fusions, sequence variants, and altered gene expression, which can be used more conveniently in clinical laboratories.
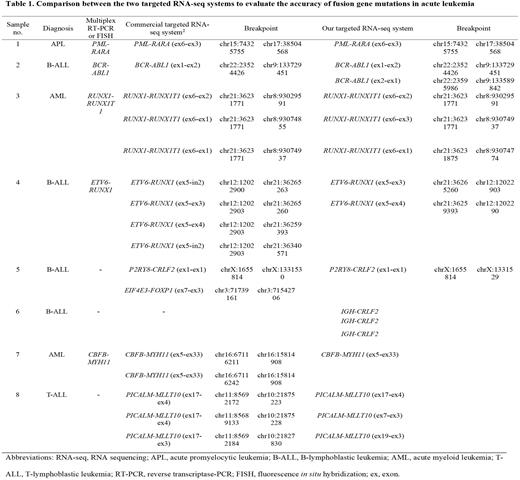
Methods: The targeted RNA-seq panel and analysis pipeline developed based on information technology with an automated reporting system and commercialized by the authors was applied to a comparative evaluation with the commercialized targeted RNA-seq panel (Archer™ FusionPlex™ Heme Panel version 2; Archer DX, Boulder, CA, USA) using samples of acute leukemia patients. A laboratory-oriented targeted RNA-seq system consisting of 84 genes was developed in consideration of the test reporting time and operational efficiency in clinical laboratories (KBB-RNAseq NGS-Leukemia-PHB; KBlueBio Inc., Hwasun, Korea). For comparison with the existing analysis system, the Archer FusionPlex Heme v2 panel (ArcherDx) based on the cDNA library for specific genes using anchored multiplex PCR was used. To evaluate the test performance of these systems, bone marrow specimens were used while diagnosing 93 patients with leukemia (15 cases of acute myeloid leukemia, 35 cases of adult B-acute lymphoid leukemia, 30 cases of childhood B-acute leukemia, and 13 cases of T-acute lymphoid leukemia).
Results: Among all 93 leukemia patients, tier 1 or tier 2 gene fusions were observed in 72 (77%) patients. Gene fusions were detected in 83% (25/30) of patients with pediatric B-acute lymphoid leukemia (B-ALL), and in 94% (33/35) of patients with adult B-ALL. In patients with acute myeloid leukemia (AML) and T-ALL, the proportions of fusion gene mutations were 53% (8/15) and 46% (6/13), respectively. For the comparative evaluation, 4 cases of B-ALL, 2 cases of AML, and 1 case of acute promyelocytic leukemia and T-ALL were used. The results of the two analysis systems were consistent in 7 of the 8 comparatively evaluated patients, but an IGH-CRLF2 fusion in a B-ALL case was detected only with the panel developed by the authors. It is known that the CRLF2 gene is a causative gene in half of the Philadelphia chromosome (Ph)-ALL cases, which account for 20-25% of all B-ALL cases and are classified as a new type of ALL showing a very poor prognosis.
Conclusions: Our targeted RNA-seq system accurately and efficiently detected various fusion gene mutations that occur in acute leukemia. Importantly, our targeted RNAseq panel detected fusion gene mutations that cause Ph-like B-ALL more accurately than the c-ommercialized test system. This targeted RNA-seq system showed a reliable and stable performance in detecting various gene fusions occurring in blood cancers, such as leukemia, in clinical laboratories. Also, our targeted RNA-seq system was found to be very useful in diagnosing Ph-like B-ALL compared to the commercial RNA-seq panel.
Keywords: Targeted RNA-seq, Clinical Laboratory, Ph-like B-ALL, Acute leukemia
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.